Inhalt: Details on our work and our goals
Our Work
Many things happened during the last years leading to exciting progress and even more valuable interconnection between researchers working on common bunt.
We received reports during the season that the disease spread even more widely across Germany, Europe and the world. Breeding for complete resistance in adapted genetic background with high yield potential is becoming more and more important. Since we need complete resistance to this fiend, stacking of several genes together is the only durable and reliable strategy.
With is why the members of the ICBC are working together on starting to solve those problems.
From institutes over breeders to single persons every involved member is working add their share of information.
The texts below will give you an overview of the known Bt genes.
Julius Kühn Institut, BrandResist project | Claire Ferreira, Albrecht Serfling & Andreas Stahl
The goal of the BrandResist project is to find and use previously unused resistances from diverse genetic materials. To screen for susceptibility, several pathogen isolates of European origin will be tested to analyse the genotypes. To identify QTL´s genotyping and genome-wide association studies (GWAS) will be done. Using NAM populations, we can develop molecular markers for resistance QTL´s. Resistance-specific KASP markers can be derived from these, which significantly speed up the breeding of resistant varieties.
Finishing the first year of the project, we can report that the first field trial was successful and the first GWAS data are being analysed right now. The second field trial is growing slowly and will, hopefully, give us more insides and some more interesting QTL´s to look at.
Download
Bt2
Mapping Bt2 has been difficult and we finally have an idea why. So far, four different genes have been identified giving identical infection patterns in phenotyping with the used virulence races and two/three of them (Bt2 differential + Bussard (Bt_Bussard+ Bt_magnifik_5D)) even behave identically across all 44 virulences races in the PAN Europe experiment (Borgen et al 2023). This resulted in lack of power in the GWAS and the signals are still not impressive, but at least consistent between Blink, FarmCPU and MLM. Observe the lack of signal at 7A, where we have BtH from an earlier analysis.
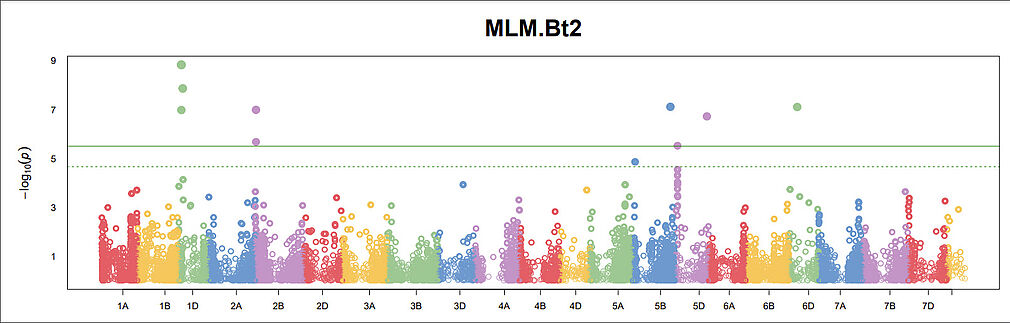
The four genes with virulence pattern identical to Bt2 are:
- The real Bt2 found in Hussar and present in the differential PI 554097 on Chromosome 1D
- Then there is BtHereward/BtH/BtQ_7A. This gene is found alone in Skagen and Inthaler and some breeding lines.
- One gene at 2B preliminary named Bt_Bussard_2B. This gene appears to be responsible for the Bt2-like resistnce in varieties such as ’Dream’, ’Paroli’ and ’Complet’.
- One gene at 5D preliminary named Bt_Magnifik_5D. This gene is found alone in a few breeding lines descending from ’Skotte’ or’Quebon’, but it is not possible to say anything definite about the phenotyping pattern.
Fine mapping of the four genes is a work in progress and the quality is still low. Due to lack of parents/offspring triplets it is not possible to identify recombination events precisely and simple haplotype comparison is not as robust.
Virulence against Bt2 is present in ~50% of all bunt races in Europe (Borgen et al 2023A). This can be explained by the fact that the three new genes found to give the same virulence pattern as the original Bt2 is unintended present in many European varieties.
Some lines have markers for multiple “Bt2” loci and we do not know how many loci/genes are actually present. Some notable examples are:
- ‘Hereward’ with the markers for Bt2, Bt_Bussard_2B and BtH
- ‘Bussard’ with Bt_Bussard_2B and Bt_Magnifik_5D
- ‘Butaro’ with Bt_Bussard_2B
- Bt_Magnifik_5D( and Bt7)
- ‘Skotte’ with Bt_Bussard_2B and Bt_Magnifik_5D
- ‘Quebon’ and ‘Format’ with BtH, Bt_Bussard_2B and Bt_Magnifik_5D.
Plenty of material exist that can be used for mapping. ‘Hereward’ is a founder in the NIAB 8 founder MAGIC population and ‘Format’ + ‘Bussard’ are founders in the BMW MAGIC population. Hussar has been used in US wheat breeding and many lines descending from it are available from genebanks.
Bt4 and Bt6
Bt4 and Bt6 are behaving identically with all virulence races so far in historic trials and in Anders, BOKU and Utah trials (100+). Both genes are at 1B and are either the same gene or tightly linked. Many historic and well characterized Bt4 containing lines were genotyped. We have a good mapping of Bt6 and studying haplotypes in an extended interval around the Bt6 interval in the Bt4 lines gave a reasonable mapping. The Bt4 and Bt6 intervals are overlapping and Bt4/Bt6 lines have identical haplotypes in the Bt6 interval.
For breeding and for the differential set there is no value in keeping both Bt4 and Bt6, and no harm.
Bt5
Dennis mapped the Bt5 to an interval on 1B (163,225,664 – 283,930,031 bp) (unpublished) and markers are available for MAS. Due to lack of marker polymorphism across the interval, we get many false positives. In our panel 30%. There is much better marker polymorphism just outside the interval and we can get the false positive rate down to 5% by adding some markers outside the interval, but at the cost of an increased false negative rate, from 5 to 10 %.
Our panel had very few lines where we had the parents as well, and it was therefore not possible to detect recombination events. We genotyped 90 lines described in the old papers this year and many of them have Bt2 in addition to Bt5. This helped improve the mapping very much.
Dennis had to resort to haplotype comparisons and this is not as solid as detecting recombination events in parents-offspring triplets. Fortunately, we have new populations in the pipeline where the parents were selected to give good marker contrast in the candidate interval.
Bt8
Bt8 was discovered in ‘Yayla 305’ (PI 178210) by Waud and Metzger (1970) and used as differential until the Goates (1996, 2012) update, where ‘M72-1250’ (PI 554120) replaced it. ‘M72-1250’ (PI 554120) inherits Bt8 from ‘7845’ (PI 173438), which was shown to have it by a test cross to ‘Yayla 305’ (PI 178210). Neither of the two sources of Bt8 have been genetically mapped in either of these varieties and it is not firmly established that ‘M72-1250’ (PI 554120) is monogenic and its resistance is identical to Bt8 in ‘Yayla 305’ (PI 178210). Also the ‘Yayla 305’ (PI 178210) Bt8 gene is possibly not monogenic. The Turkish landrace ‘6256’ (PI 178383) was shown to carry Bt8 and has served as the major source of Bt8 in especially US wheat breeding, but also in Sweden.
As a first step towards making a Starke II based Bt8 NIL, Anders has crossed Starke II and PI 554120. From this population we have genotyped and phenotyped 13 RILs. Two of these RILs are nearly NILs (98.6% identical), one with Bt8 and one without, and Dennis used them to get a few candidate intervals. All but one on 4B could be dismissed by including a few more RILs and the parents.
The phenotyping results give a clear indication that PI 554120 has two genes. We have no good mapping of the second gene.
From testing the 4B marker block in the entire panel, we conclude that it looks promising to be the dominating gene Bt8 from PI554120.There are approximately 200 Bt8 lines in the field in 2024. Approximately half descending from PI 554120 and half from Magnifik (and therefore from PI 178383 via Stava), which is another potential assigned carrier of Bt8.
We have no solid, or even semi solid, information on the original Bt8 described in ‘Yayla 305’ yet. The genebank accession of Yayla 305 seems heterogeneous which complicates the search for Bt8 in this original source.
Bt9
Bt9 has been mapped to 2B (Steffan et al 2017) and a refined mapping was presented at the Tulln Workshop 2023. (Christensen and Borgen 2023B)
Later research demonstrated that two of the markers published were wrong. They are not at 6D and have been removed (Thanks Almuth!). The interval has not changed but the new markers for MAS are:
Table 2 New Markers for mapping Bt9
| Kukri_rep_c107605_164 | T |
| wsnp_CAP8_rep_c4586_2232878 | C |
| wsnp_CAP7_c1735_859875 | G |
| wsnp_CAP7_c1735_859744 | T |
Bt10 and BtZ
Bt10 and BtZ are two genes with a lot in common and may even be identical. Phenotypically they are identical, and they are both present at Chromosome 6D. However, the mapping is not 100% identical.
Bt10 has been mapped to 6D (Laroche et al 2000, Menzies et al 2006) and a refined mapping was presented at the Tulln Workshop 2023. (Christensen and Borgen 2023C)
BtZ has been also been mapped to 6D as presented at the Tulln Workshop 2023. (Chrisenten and Borgen 2023E)
After the Tulln workshop, Dennis went back to the BtZ mapping and redid it getting a slightly different result.
Anders has made a small RIL population from the cross Starke II x Inna. Lines are called NIL-Z because they are the first step towards making a Starke II based BtZ NIL.
Table 3 Markers to potentially detect
| BtZ RAC875_rep_c118305_446 | T |
| wBS00065960_51 | C |
| Kukri_c73802_205 | A |
Orange markers are flanking the QTL interval potentially including BtZ. Green marker may be used for MAS, but monomorphic in this population There are two RefSeq High Confidence genes in that interval
Table 4 RefSeq High Confidence genes
| Chromosome | Phys Pos Min | Phys Pos Max | Gene | Function |
|---|---|---|---|---|
| Chr6D | 4363458 | 4366232 | TraesCS6D03G0022800 | Receptor-like protein kinase |
| Chr6D | 4533591 | 4537917 | TraesCS6D03G0023800 | F-box family protein |
Bt10 and BtZ behaves identically with all virulence races used in the PAN Europe trial (Borgen et al 2023A) and in all previous trials by Anders and they map to near identical intervals at 6D. The original BtZ interval presented last year in Tulln overlapped with the Bt10 interval, but the new one presented here does not. Dennis is convinced that the new Bt10 interval is inaccurate and that BtZ = Bt10 = TraesCS6D03G0022800. Phenotyping 75 Saatsucht Donau lines from a Bt10 x BtZ cross we found 1-2 lines with a single or a few infected heads. It remains to be investigated whether this is true segregation or an error.
BtZ is supposed to be present in Zarya as a Thinopyrum intermedium introgression, inherited from AG.IN via PPG-599 and Lutescens.126-65.

It seems a bit unlikely that such an introgression should contain Bt10 and end up at 6D. To investigate that, Mironovskaya 808 and PPG-599 will be phenotyped. Lutescens-126-65 and Smes-Sortov are available from the Vavilov Institute (https://www.wheat-gateway.org.uk/search.php?send=1&per=50&search=sortov&bunt_a=1&genes=1&simple=1). Thank you to Andrew Forbes for the information!
Bt11 and Dimenit Genes
Lunzer et al 2023 mapped a number of loci/genes in the Bt11 differential line ‘M82-2123’ (PI 554119) and in ‘Dimenit’ (PI 166910).
Four biparental mapping populations were used: M82 2123 x Mulan, Rainer x Dimenit, Dimenit x Rainer and Dimenit x Lukullus. Three loci at 4BS, 4BL and 6DL were mapped in M82-2123 and a fourth locus at 2A was segregating in that population. In the Rainer x Dimenit and Lukullus x Dimenit populations two loci at 4BS and 6DL were mapped. Dimenit was the donor. It turned out that Dimenit was not homogenous and in the Dimenit x Rainer population, and three (or four) additional loci at 1A, 1B and 7B were found. It is uncertain whether the 4BL locus was identified in this population as the 4B QTL spanned the entire chromosome.
All phenotyping and genotyping data were made public – excellent! Thank you BOKU!
Dennis (unpubllished) did a detailed analysis of the data and found that our Bt3 markers detected the gene at 1A, and the Bt6 markers detected the gene at 1B. The Bt7 markers were also validated. Bt7 is not very effective against the virulence used in the BOKU study and was therefore not detected, but still this gene was segregating in the population.
During the initial population validation, Dennis discovered that the Dimenit selection with the extra genes was identical to the one we have later genotyped. Dennis also discovered lines having the wrong parents and lack of inbreeding/selection, especially in the Dimenit x Lukullus population.
The detailed analysis identified recombination events that could be used to refine the intervals from the QTL mapping, and in most cases gave much smaller intervals. However this was not possible for the 4BL and 7B loci as these were masked by the other loci in most lines and not very effective against the used inocula.
The 6D locus gives immunity to both inocula used by Lunzer et al (2023), but interestingly, the 4BS locus was highly resistant to the BOKU “house keeping” inoculum, but the “aggressive” inoculum used in 2022 was able to give infections in the 12-24 % range.
Unfortunately we have not managed to get lines from the mapping populations with each gene isolated, into the field trial of 2024, except for a few exceptions; U11.15 with the 6D locus and U11.50 with the 7Blocus. From the 2022 trial, we know that breeding lines from Anders with the 6D locus alone is fully resistant to all eight virulence races in Anders´ core set.
Getting lines with 4BS and 4BL alone in the 2024 trial was a high priority, but we did not succeed in finding such lines.
Two lines have recombination in the Bt3 interval (U11.114 and U11.84) but they also have the 6D gene that is masking the presence or absence. Perhaps we can find a virulence race virulent on 6D but not on Bt3 to lift the mask. We could also cross the lines to susceptible varieties to test if Bt3 segregates. Any volunteers? ; )
A big question is whether the 6D QTL in Dimenit may be identical to the Bt9 gene. We have this evidence so far:
- Abdallah et al. (1984) found Bt7, Bt9 and Bt11 in Dimenit
- Bt9 and Dimenit 6D both are at 6DL
- The haplotypes of Bt9/Bt11 lines are very different around the mapped intervals
- There is no overlapping of Dennis’s intervals for Bt9 and Dimenit 6D so far
Additional evidence can be collected by phenotyping 6D lines with the common bunt races L-21/T35 (Goates 2012) which are virulent to Bt9, but not to Bt11. Any volunteers?
Based on our analysis, we preliminarily suggest replacing Bt11 by two new assigned Bt gene numbers and also exchange the line in the differential set by the separate QTL carriers. Nevertheless, a bit of validation work has to be done still.
Bt12 and other PI 119333 Genes
Bt12 was mapped by Muellner et al (2020) to 7DS in the interval 6.5 – 10.8 Mbp in RefSeq 1.0 positions. Chromosome 4B was found to contribute some resistance too. The 7DS interval was found not to be 100% linked to Bt12, meaning that it is a signal only and actually an exclusion interval.
BOKU kindly gave Dennis access to genotypic and phenotypic data, which he analysed.
Anders phenotyped 37 lines from the mapping populations with 7-9 virulence races and we found that two genes at 7D and two at 4B could explain the infection patterns.
To get a mapping at 4B, Dennis analysed markers across the chromosome and found 18 recombination events, giving 19 intervals. Interval 1 at 1,306,080 – 15,855,852 bp was found to be harbour some resistance gene(s). Intervals 2-14 were not related to the presence of resistance and interval 15-19 at 648,869,446 – 670,633,612 bp (end of chromosome) again provided resistance.
In this table, we see the infection patterns for the four genes and for their combinations. It was very difficult to figure this out based on the patterns from the 37 lines only and there may be errors in it. There may also be inaccuracies and errors in the phenotyping and the table should not be taken as the final word.
Table 5 Infection pattern from Bt4, Bt12 and their combinations Vr0
| Vr-2 | Vr-3 | Vr-5 | Vr-10 | Vr-13 | Vr-Z | Vr-DOT | Vr-2+Iran | |
| Bt4B_1 | Bt4B_1 | Bt4B_1 | ||||||
| Bt4B_2 | Bt4B_2 | Bt4B_2 | Bt4B_2 | |||||
| Bt12A | Bt12A | Bt12A | Bt12A | |||||
| Bt12AB | Bt12B | Bt12B | Bt12B | |||||
| Bt4B_1+Bt4B_2 | Bt4B_1+Bt4B_2 | |||||||
| Bt12A+Bt12B | Bt12A+Bt12B | |||||||
| Bt4B_1+Bt12A | Bt4B_1+Bt12A | |||||||
| Bt4B_2+Bt12A | ||||||||
| Bt4B_1+Bt12B | Bt4B_1+Bt12B | Bt4B_1+Bt12B | ||||||
| Bt4B_2+Bt12B | Bt4B_2+Bt12B | Bt4B_2+Bt12B | ||||||
| Bt4B_1+Bt4B_2+Bt12A | ||||||||
| Bt4B_1+Bt4B_2+Bt12B | Bt4B_1+Bt4B_2+Bt12B |
We see that the perceived strength of “Bt12” comes from the good combining ability of the four genes in PI 119333.

